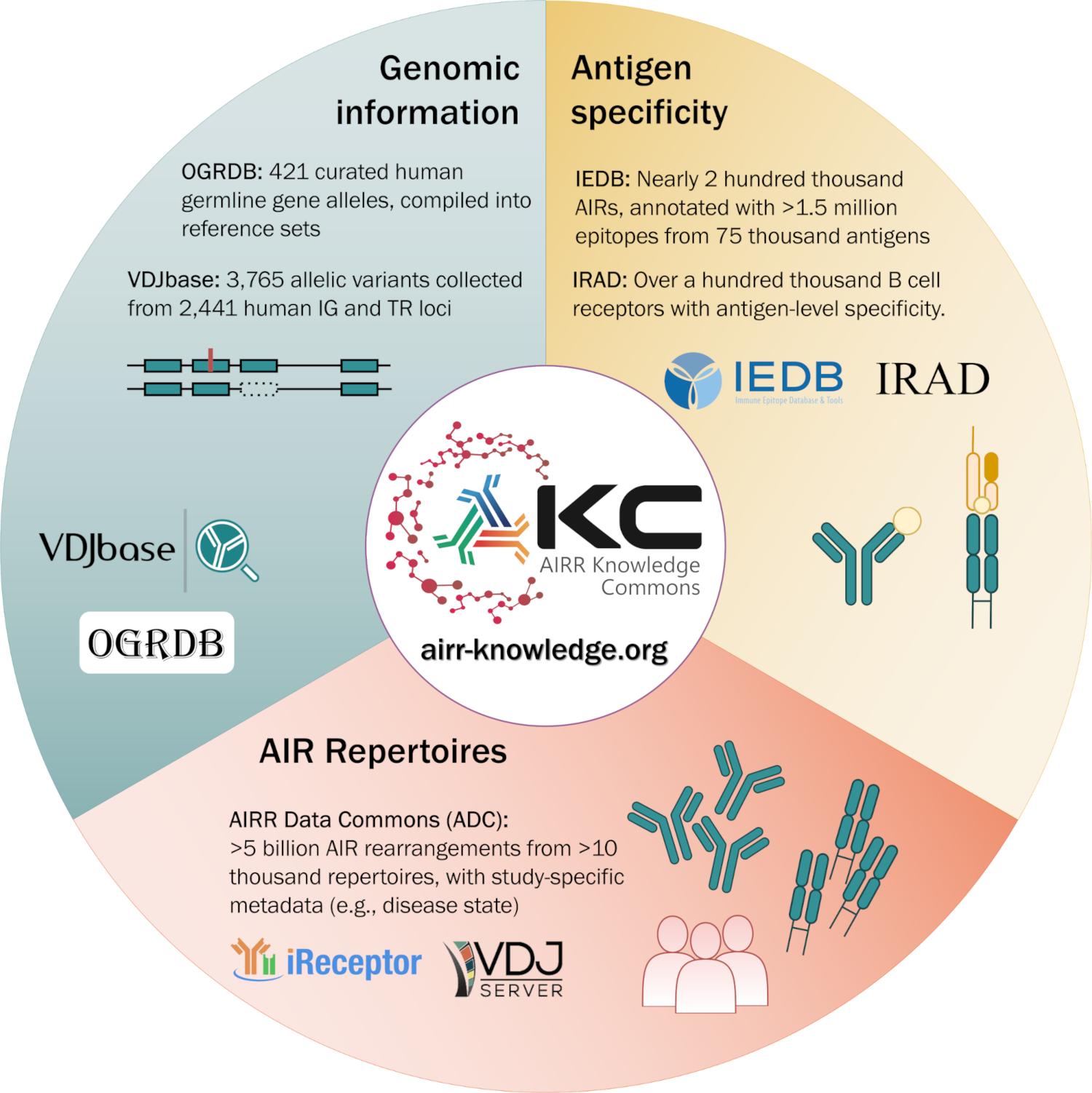
A bridge between immune receptor databases
The AKC is being established by merging data from existing, community-backed repositories and applying existing and novel, cross-cutting knowledge-generation algorithms to the integrated data. The AKC currently includes:
- Repertoire data from the AIRR Data Commons, which contains >10,000 repertoires with billions of AIRs
- AIR germline allele, genotype, haplotype, and population genetic data from the Open Germline Receptor Database and VDJbase
- AIR specificity data from the Immune Epitope Database and Immune Receptor Antigen Database.
The AKC defines a common data model to harmonize data from different repositories, allowing users to query and perform analysis across integrated data.
Data submission
The AKC sources its data from its different component repositories. Please submit your data to the appropriate repository, for integration into the AKC.
- For AIRR-seq studies, please read the AIRR community data submission guidelines.
- AIRR-seq study data may be deposited in VDJServer.
- Alternatively, you can set up your own AIRR-compliant data repository by creating an iReceptor turnkey. This is recommended for very large datasets.
- Previously undocumented immune receptor alleles in AIRR-Seq repertoires can be submitted to OGRDB. See the OGRDB data submission guide.
- Experiments determining TCRs or BCRs binding to specific epitopes can be submitted to the IEDB. See IEDB data submission.
- Experiments determining antigen specificity without specific epitope information should be stored in IRAD. IRAD sources its data from GenBank. To request that your sequences depositied in GenBank be scheduled for curation, please contact Krishna.Roskin@cchmc.org.
If you are unsure where to submit, feel free to reach out to airr-knowledge@utsouthwestern.edu
